Amplicon Sequencing Prices. as low as $45/assay for V4 16s sequencing
| Sequencing Platform | Read Length /Assay Length | Pricing |
| Ion S5 XL** | 300bp 515F bacterial + archaeal or ITS1-2 fungal diversity | For nominal 15-20,000 reads/assay: |
| $60/assay (any size project) | ||
| Ion S5 XL (EXT)** | >500bp bacterial, archaeal, | Same Pricing as Illumina MiSeq |
| or ITS fungal diversity assays | (No Additional Library Fee) | |
| 20,000 sequence diversity assays starting at: | ||
| $60 /assay (>150 assays) | ||
| $65/assay (100-150 assays) | ||
| Illumina MiSeq | 2×300 PE | $70/assay (50-100 assays) |
| $75/assay (20-50 assays) | ||
| $80/assay (1-20 assays) | ||
| For projects < 10 samples a $100 library fee is added | ||
| Low Coverage (< 500 Seqs): $50/assay | ||
| ~5,000 Seqs: $90/assay | ||
| PacBio Sequel | Long Read Amplicon Sequencing | ~10,000: $125/assay |
| (16s, 18s, ITS, and Custom) | ~20,000: $200/assay | |
| ~40,000: $340/assay | ||
| Custom Coverage available; Contact us for pricing | ||
| For projects < 10 samples a $150 library fee is added & $20/sample PCR replication fee |
MR DNA has all the major sequencing platforms and we have a large selection of amplicon sequencing programs .. Everything can be customized to the needs of the customer, only limited by the capabilities of the sequencing technologies.
Any amplicons (bTEFAP® services) such as 16s, 18s, ITS, functional or custom assays.. if you have an amplicon with custom primers we can help sequence it.
Illumina miseq and hiseq amplicons
examples for any of our hundreds of inhouse assays
2x300bp PE illumina 20,000 sequence diversity assays
$80/assay 1-20 assays (note: for projects < 10 assays per library, a $100 library fee is added),
$75 for 20-50 assays,
$70 for 50-100 assays,
$65 for 100-150 assays,
$60 for > 150 assays.
additional discounts for very large projects also
Ion S5 XL (Ion Torrent) amplicons**
All Ion S5 XL projects are being migrated to the Illumina MiSeq and Illumina NovaSeq Sequencing Platforms
All economy sequencing projects previously completed using the Ion S5 will now be completed using the Illumina MiSeq and NovaSeq at no additional cost.
- The same 515F-806R and ITS1-2 primers will continue to be made available for use on all Illumina sequencing platforms
- Contact us to discuss additional Illumina specific primer sets and pricing options
300bp 515F bacterial + archaeal or ITS1-2 fungal diversity Ion S5 assays with nominal 15-20,000 reads/assay for $60/assay (any size project)
>400bp bacterial, archaeal, or ITS fungal diversity assays with nominal 15-20,000 reads/assay starting at $85/assay and continues to follow the Illumina MiSeq pricing scale above.
PAC BIO SEQUEL:
Sequel long read amplicon sequencing for 16s, 18s, ITS and custom amplicons
MR DNA now accepts any size project (projects with < 10 samples do have a $125 indexing fee and a $20/sample additional PCR replication fee.)
Academic and Government Pricing
$90 per sample for 5,000 sequences per assay,
$125 per sample for 10,000 sequences per assay
$200 per sample for 20,000 sequences per assay
$340 per sample for 40,000 sequences per assay
**Custom assays from 700bp – 3000bp or larger of course have a barcoding fee that is typically $15/barcode
DNA Extraction / RNA Extraction Price:
–DNA Extraction starting at $25 / sample (depending on service requested and sample type submitted)
–RNA Extraction starting between $30 – $70 / sample (depending on service requested and sample type submitted)
| 16s rRNA Sequencing Primer List | |
| Updated Earth Microbiome Project (EMP) 16s v4 | |
| 515F | GTGYCAGCMGCCGCGGTAA |
| 806R | GGACTACNVGGGTWTCTAAT |
| Original Earth Microbiome Project (EMP) 16s v4 | |
| 515F | GTGCCAGCMGCCGCGGTAA |
| 806R | GGACTACHVGGGTWTCTAAT |
| Other Common 16s rRNA Primers | |
| 515F | GTGYCAGCMGCCGCGGTAA |
| 926R | CCGYCAATTYMTTTRAGTTT |
| 909R | CCCCGYCAATTCMTTTRAGT |
| archaea 349F | GYGCASCAGKCGMGAAW |
| A344F | AYGGGGYGCASCAGGSG |
| archaea 806R | GGACTACVSGGGTATCTAAT |
| arch21F | TTCCGGTTGATCCYGCCGGA |
| arch519R | TTACCGCGGCGGCTG |
| arch1059R | GCCATGCACCWCCTCT |
| arc344F | ACGGGGYGCAGCAGGCGCGA |
| arch915R | GTGCTCCCCCGCCAATTCCT |
| 27F | AGRGTTTGATCMTGGCTCAG |
| 519Rmod | GTNTTACNGCGGCKGCTG |
| 519Rmodbio | GWATTACCGCGGCKGCTG |
| 1492R | GGGTTACCTTGTTACGACTT |
| 338R | AGTGCTGCCTCCCGTAGGAGT |
| 28F | GAGTTTGATCNTGGCTCAG |
| 519R | GTNTTACNGCGGCKGCTG |
| 341F | CCTACGGGNGGCWGCAG |
| 785R | GACTACHVGGGTATCTAATCC |
| 805R | GACTACNVGGGTATCTAATCC |
| 799F | ACCMGGATTAGATACCCKG |
| 1193R | CRTCCMCACCTTCCTC |
| a799wF | AMCVGGATTAGATACCCBG |
| new1193R | ACGTCATCCCCACCTTCC |
| 16com1F | CAGCAGCCGCGGTAATAC |
| 16com2R | CCGTCAATTCCTTTGAGTTT |
| 926F | AAACTYAAAKGAATTGACGG |
| 1394R | ACGGGCGGTGTGTRC |
| Tx9F | GGATTAGAWACCCBGGTAGTC |
| 1391R | GACGGGCRGTGWGTRCA |
| 1100F | YAACGAGCGCAACCC |
| 1492R | GGGTTACCTTGTTACGACTT |
| rambacV3F | CCTACGGGAGGCAGCAG |
| rambacV4R | GGACTACHVGGGTWTCTAAT |
| 104F | GGCGVACGGGTGAGTAA |
| 530R | CCGCNGCNGCTGGCAC |
| 530F | GTGCCAGCMGCNGCGG |
| bac926R | CCGTCAATTYYTTTRAGTTT |
| 1100R | GGGTTNCGNTCGTTR |
| 18s rRNA Sequencing Primer List | |
| EukV4F | CCAGCASCYGCGGTAATTCC |
| EukV4R | ACTTTCGTTCTTGATYRA |
| ionesV4R | ACTTTCGTTCTTGA |
| zigEukV4R | ACTTTCGTTCTTGATYRATGA |
| euk1391F | GTACACACCGCCCGTC |
| EukB-Rev | TGATCCTTCTGCAGGTTCACCTAC |
| Euk7F | AACCTGGTTGATCCTGCCAGT |
| Euk570R | GCTATTGGAGCTGGAATTAC |
| uni18sF | AGGGCAAKYCTGGTGCCAGC |
| uni18sR | GRCGGTATCTRATCGYCTT |
| nem18sF | CGATCAGATACCGCCCTAG |
| nem18sR | TACAAAGGGCAGGGACGTAAT |
| paraOxyF | GCYGCGGTAATWCCAGCTCT |
| paraoxyR | TGCNCTTCCGTCAATTYCTT |
| 1080F | GGGRAACTTACCAGGTCC |
| 1578R | GTGATRWGRTTTACTTRT |
| SSU316F | GCTTTCGWTGGTAGTGTATT |
| 758R | CAACTGTCTCTATKAAYCG |
| AML1 | ATCAACTTTCGATGGTAGGATAGA |
| AML2 | GAACCCAAACACTTTGGTTTCC |
| wanda | CAGCCGCGGTAATTCCAGCT |
| 960F | GGCTTAATTTGACTCAACRCG |
| 1200R | GGGCATCACAGACCTG |
| 1560F | TGGTGCATGGCCGTTCTTAGT |
| 2035R | CATCTAAGGGCATCACAGACC |
| Euk60F | GAAACTGCGAATGGCTCATT |
| Euk515R | ACCAGACTTGCCCTCC |
| Euk516F | GGAGGGCAAGTCTGGT |
| Euk1055R | CGGCCATGCACCACC |
| eukss18F | CACCAGGTTGATTCTGCC |
| eukss530R | GTGCCAGCMGCCGCGG |
| Euk528F | CCGCGGTAATTCCAGCTC |
| EukR18R | CGTTATCGGAATTAACCAGAC |
| SSUF04 | GCTTGTAAAGATTAAGCC |
| SSUR22 | GCCTGCTGCCTTCCTTGGA |
| ITS Sequencing Primer List | |
| ITS1F | CTTGGTCATTTAGAGGAAGTAA |
| ITS2R | GCTGCGTTCTTCATCGATGC |
| ITS4R | TCCTCCGCTTATTGATATGC |
| ITS3F | GCATCGATGAAGAACGCAGC |
| ITS3kyo2F | GATGAAGAACGYAGYRAA |
| ITS5F | GGAAGTAAAAGTCGTAACAAGG |
| ITS7 | GTGARTCATCGAATCTTTG |
| ITS9 | GAACGCAGCRAANNGYGA |
| ITS6pyth | GAAGGTGAAGTCGTAACAAGG |
| ITS7Rpyth | AGCGTTCTTCATCGATGTGC |
| gITS7F | GTGARTCATCGARTCTTTG |
| ITS4ngsR | TTCCTSCGCTTATTGATATGC |
| ramITS1ooF | CGGAAGGATCATTACCAC |
| ramITS58ooR | AGCCTAGACATCCACTGCTG |
| ENDONTSF | AAGGTCTCCGTAGGTGAAC |
| ENDONTSR | GTATCCCTACCTGATCCGAG |
| its58funFbar1 | AACTTTYRRCAAYGGATCWCT |
| its4funR | AGCCTCCGCTTATTGATATGCTTAART |
| symbiITS1 | GAATTGCAGAACTCCGTG |
| symbiITS2 | GGATCCATATGCTTAAGTTCAGCGGGT |
| traceITS1ooF | GGAAGGATCATTACCACAC |
Why Should I Choose the 16s Sequencing Service?
Prokaryotes today are divided into two domains, Archaea and Bacteria. These two domains are of particular interest in areas of research including:
- Soil Ecology
- Gastroenterology
- Medical microbiology
- Food Science
- etc.
The initial objective of the many studies within these fields among others is often the same; identify which microbes are present, or more importantly, which are absent. The 16s rRNA gene is an excellent sequencing target in order to complete such studies. There are nine hypervariable regions found in the 16s rRNA gene, and each of these regions is flanked by a highly conserved region. Our in-house 16s rRNA sequencing primer pairs are specifically designed to target these flanking conserved regions thereby allowing us at MR DNA to perform PCR amplification and DNA sequencing of your submitted microbial samples. Of the nine hypervariable regions found in the 16s rRNA gene, some regions may be better suited to complete certain phylogenetic studies over others. Feel free to consult our experts at MR DNA is order to determine which 16s rRNA primer pair is best suited to meet your sequencing needs.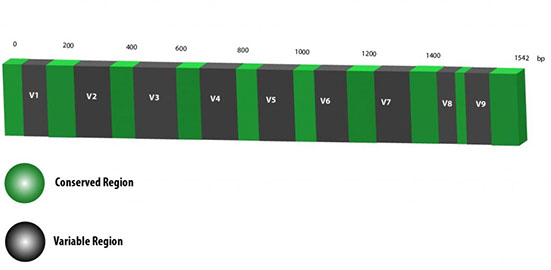
The already cost effective method of 16s rRNA sequencing continues to reduce in cost as sequencing technology continues to advance. By utilizing the technology made available by next-generation sequencing platforms, we are able to generate the necessary data required to complete these 16s rRNA phylogenetic studies in a much more time efficient and cost-effective manner. For more information concerning our 16s rRNA sequencing capabilities, feel free to Contact us.
What is 16s rRNA Sequencing?
16s rRNA sequencing has become one of the leading methods for phylogenetic studies. The popularization of 16s sequencing methods has been due in large part to the wide availability of PCR and Next-generation sequencing facilities, such as MRDNA. But what is 16s rRNA sequencing? And why should you choose 16s sequencing methods over other DNA sequencing methods?
16s rRNA sequencing refers to sequencing the 16s rRNA gene that codes for the small subunit (SSU) of the ribosome found in prokaryotes such as Bacteria and Archaea. There are several factors that make the 16s rRNA gene the perfect target to complete your taxonomy or phylogeny studies.
- Because the 16s gene codes for the SSU of the prokaryotic ribosome, researchers can rely on the fact that the their target gene will be present in every cell.
- The 16s gene contains both highly conserved regions as well as hypervariable regions.
- The presence of the highly conserved regions allow researchers to design primer pairs that will accurately and reliably amplify the 16s hypervariable region of their choice.
- The presence of the hypervariable regions affords researchers the ability to differentiate between closely related genera or species detected in their samples.
- The overall size of the 16s rRNA gene is relatively short. ~1500bp. While sequencing the entire 16s gene is difficult due to read length restrictions of many NGS platforms, sequencing one or more hypervariable regions is relatively quick and affordable.
- Two of our most requested assays for 16s rRNA sequencing are 27F-519R (V1-V3 region) and 515F-806R (V4 region).
- For questions regarding pricing feel free to contact us or visit our 16 ribosomal sequencing page.
Often times, researchers will have some confusion regarding the differences between 16s metagenomic sequencing methods and shotgun metagenomic sequencing methods. In short, shotgun metagenome sequencing is aptly named due to the fact that the goal of this DNA sequencing method is to sequence all genes from all organisms in a given sample. Whereas in the case of 16s metagenome sequencing, the goal is to sequence the 16s rRNA gene specifically.
Related Research
Periodontitis is caused by dysbiotic subgingival bacterial communities that may lead to increased bacterial invasion into gingival tissues. Although shifts in community structures associated with transition from health to periodontitis have been well characterized, the nature of bacteria present within the gingival tissue of periodontal lesions is not known. To characterize microbiota within tissues of periodontal lesions and compare them with plaque microbiota, gingival tissues and subgingival plaques were obtained from 7 patients with chronic periodontitis. A sequencing analysis of the 16S rRNA gene revealed that species richness and diversity were not significantly different between the 2 groups. However, intersubject variability of intratissue communities was smaller than that of plaque communities. In addition, when compared with the plaque communities, intratissue communities were characterized by decreased abundance of Firmicutes and increased abundance of Fusobacteria and Chloroflexi. In particular, Fusobacterium nucleatum and Porphyromonas gingivalis were highly enriched within the tissue, composing 15% to 40% of the total bacteria. Furthermore, biofilms, as visualized by alcian blue staining and atomic force microscopy, were observed within the tissue where the degradation of connective tissue fibers was prominent. In conclusion, very complex bacterial communities exist in the form of biofilms within the gingival tissue of periodontal lesions, which potentially serve as a reservoir for persistent infection. This novel finding may prompt new research on therapeutic strategies to treat periodontitis.
Baek K, Ji S, Choi Y. Complex Intratissue Microbiota Forms Biofilms in Periodontal Lesions. J Dent Res. 2017;:22034517732754.
In The News
Microbial genome sequencing is helping to improve our understanding of human health, disease, and microbial evolution. The human body contains trillions of cells with a variety of microbes that play a critical role in human health and disease, but the area of mechanism remains a mystery. Microbes are not only present in the human body; they are everywhere e.g. human or animal guts, homes, plants, oceans, and soil. Microbial research has gone under-appreciated for a long time, but with the help of next-generation sequencing (NGS), scientists are now investigating this vast microbial world. Multiple studies have been published in the last 5-10 years examining the microbial communities that exist inside our bodies and how these microbiomes can be influenced by the environment. The microbiome of the human gut can be rapidly and accurately cataloged by shotgun metagenomic sequencing via the Illumina NovaSeq 6000 System. Fecal samples, which have an abundant amount of microbes present (making it ideal to extract microbial DNA for genome sequencing), are a great candidate to help determine what is happening in the human GI tract. With NGS systems like the Illumina NovaSeq 6000, the scientific community is now able to generate gigabytes of data per sample. Many researchers today are excited about the value of microbiome sequencing and a number of clinicians believe it will become a routine part of health care, much like a blood draw.
But don’t limit microbiome sequencing to doctor’s offices and research laboratories. Direct-to-consumer tests are taking advantage of microbiome sequencing as well. One-time users are able to get an accurate snapshot of their gut health, and for the more avid citizen scientist, there are also time-series sampling options so that one can follow changes in their gut microbiome over time. So, whether the goal is to track the effect of a new diet or the efficacy of your favorite probiotic, or maybe you’re just curious…the microbial genomic data generated by microbiome sequencing can help us all to better understand our body and health, and track changes in the gut over time. NGS is continually helping us to understand the genetic blueprints of organisms within communities and obtain genome sequences from more complex environments like the human gut. Services like these were at one time too expensive for some laboratories and consumers, but with high-throughput sequencers like the Illumina NovaSeq, a comprehensive view of complex microbial environments is now available to everyone for a fraction of the cost.
1. Makki K, Deehan EC, Walter J, et al. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe. 2018; 23:705—715.
2. Holscher HD. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes. 2017; 8:172—184.
3. De Vadder F, Grasset E, Mannerås Holm L, et al. Gut microbiota regulates maturation of the adult enteric nervous system via enteric serotonin networks. Proc Natl Acad Sci U S A. 2018; 115:6458—6463.
4. Cheung SG, Goldenthal AR, Uhlemann AC, et al. Systematic Review of Gut Microbiota and Major Depression. Front in Psychiatry. 2019; 10:34.
This is an example page. It’s different from a blog post because it will stay in one place and will show up in your site navigation (in most themes). Most people start with an About page that introduces them to potential site visitors. It might say something like this:
Hi there! I’m a bike messenger by day, aspiring actor by night, and this is my website. I live in Los Angeles, have a great dog named Jack, and I like piña coladas. (And gettin’ caught in the rain.)
…or something like this:
The XYZ Doohickey Company was founded in 1971, and has been providing quality doohickeys to the public ever since. Located in Gotham City, XYZ employs over 2,000 people and does all kinds of awesome things for the Gotham community.
As a new WordPress user, you should go to your dashboard to delete this page and create new pages for your content. Have fun!